UNIT ____: Nucleic Acids Name: _____________________
Essential Idea(s):
The structure of DNA allows efficient storage of genetic information.
IB Assessment Statements
7.1.S1 | Analysis of results of the Hershey and Chase experiment providing evidence that DNA is the genetic material. - State the experimental question being tested in the Hershey and Chase experiment.
- Explain the procedure of the Hershey and Chase experiment.
- Explain how the results of the Hershey and Chase experiment supported the notion of nucleic acids as the genetic material.
|
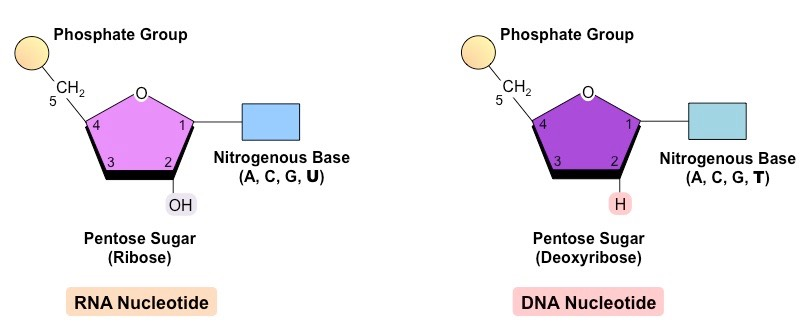
2.6.U1 | The nucleic acids DNA and RNA are polymers of nucleotides. - State the two types of nucleic acid.
- Outline the parts of a nucleotide.
- Identify and label carbons by number (for example, C1, C2, C3) on a nucleotide drawing.
- Explain how nucleotides can connect to form a nucleic acid polymer.
- State the names of the nitrogenous bases found in DNA and RNA.
- Identify nitrogenous bases as either a pyrimidine or purine.
- State the complementary base pairing rules.
|
2.6.S1 | Drawing simple diagrams of the structure of single nucleotides of DNA and RNA, using circles, pentagons, and rectangles to represent phosphates, pentoses and bases. - Draw the basic structure of a single nucleotide (using circle, pentagon and rectangle).
- Draw a simple diagram of the structure of RNA.
- Draw a simple diagram of the structure of DNA.
- Identify and label the 5’ and 3’ ends on a DNA or RNA diagram
|
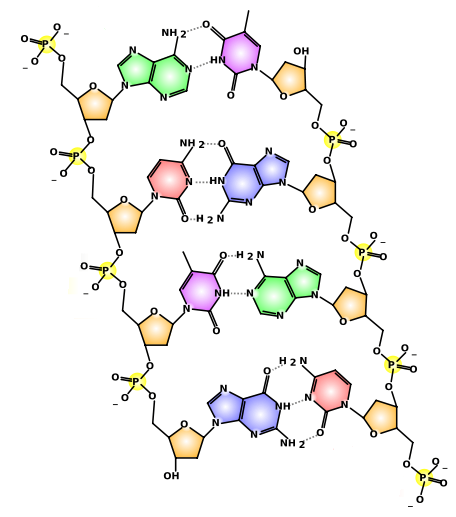
2.6.U3 | DNA is a double helix made of two antiparallel strands of nucleotides linked by hydrogen bonding between complementary base pairs. - Define antiparallel in relation to DNA structure.
- Outline the formation of a DNA double helix by hydrogen bonding between nitrogenous bases.
- Identify the four bases of DNA based on the numbers of rings (purines or pyrimidines) and the number of hydrogen bonds it can form.
- State the number of nitrogenous bases per complete turn of the DNA double helix.
|
7.1.A1 | Rosalind Franklin and Maurice Wilkins’ investigation of DNA structures by X-ray. - Outline the process of X-ray diffraction.
- Outline the deductions about DNA structure made from the X-ray diffraction pattern.
|
7.1.NOS | Making careful observations- Rosalind Franklin’s X-ray diffraction provided crucial evidence that DNA is a double helix. - Describe Rosalind Franklin’s role in the elucidation of the structure of DNA.
|
2.6.A1 | Crick and Watson’s elucidation of the structure of DNA using model making. - Outline the role of Chargaff, Watson, Crick, Franklin and Wilkins in the discovery of DNA structure.
- Explain how Watson and Crick used model building to determine the structure of DNA.
|
2.6.NOS | Using models as representation of the real world- Crick and Watson used model making to discover the structure of DNA. - List types of models used in science.
- State a common feature of models in science.
- List ways in which models are different from the structure or process it represents.
|
2.6.U2 |
DNA differs from RNA in the number of strands present, the base composition and the type of pentose. - Compare the structure of DNA and RNA.
|
3.5.U1 | Gel electrophoresis is used to separate proteins or fragments of DNA according to size. - Match restriction enzyme names to the bacteria in which they are naturally found.
- Describe the role of restriction enzymes in nature and in biotechnology applications.
- Contrast sticky vs. blunt ends.
- Identify a restriction site as either leaving sticky or blunt ends.
- Determine the number and size of DNA fragments after being exposed to restriction enzymes (both linear and plasmid DNA).
- Demonstrate accurate use of a micropipette.
- Explain the function and purpose of DNA electrophoresis.
- Describe how and why DNA fragments separate during electrophoresis.
- Outline the functions of the buffer, marker and loading dye in DNA electrophoresis.
|
3.5.U3 | DNA profiling involves comparison of DNA. - Outline the process of DNA profiling.
|
7.1.A2 | Tandem repeats are used in DNA profiling. - Define VNTR.
- Explain why VNTR are used in DNA profiling.
|
3.5.A1 | Use of DNA profiling in paternity and forensic investigations. - List example sources of DNA that can be used in DNA profiling.
|
3.5.S2 | Analysis of examples of DNA profiles. - Analyze a DNA profile to determine relatedness or forensic guilt.
|
The Hershey – Chase Experiment
EXPERIMENTAL QUESTION:
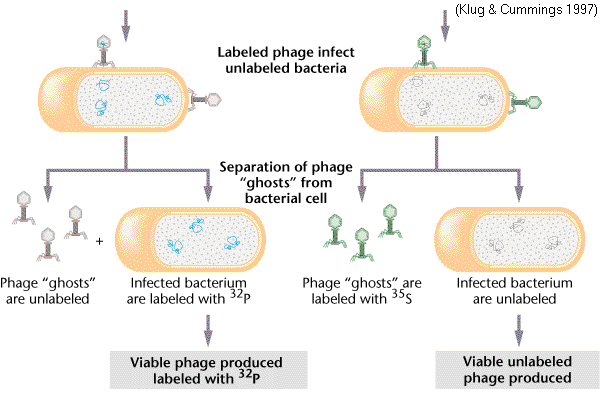
CONCLUSION:
Nucleic Acids and Nucleotides
- nucleic acids are macromolecules that:
- nucleic acids are chains of:

- nucleotides are composed of:
- There are two types of pentose sugars that are found in nucleotides:
DNA Structure
5’
| 3’ |
Backbone
| Antiparallel |
Purine
| Pyrimidine |
Hydrogen bond
| Covalent Bond |
Double Helix
| Complementary Strands |
Key Scientists in the Discovery of DNA Structure
Erwin Chargaff |
|
Rosalind Franklin |
|
Maurice Wilkins |
|
James Watson |
|
Francis Crick |
|
RNA VS DNA
| DNA | RNA |
Type of Sugar |
|
|
Number of Strands |
|
|
Structure of Sugar (drawn) |
|
|
Name of Bases |
|
|
Location(s) in the Cell |
|
|
Primary function |
|
|
Restriction Enzymes

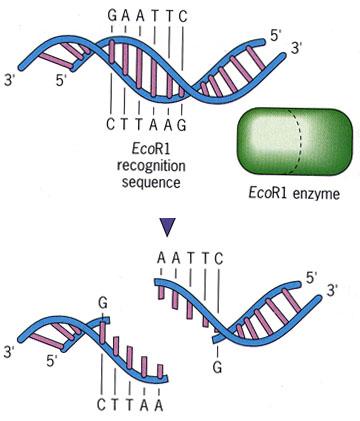
- Restriction enzymes are “molecular scissors” that ___________________________
- Restriction enzymes are isolated from __________________ for use in biotechnology research.
- The function of restriction enzymes in bacterial cells is to cut apart foreign DNA molecules (i.e. from ____________________________________)
- Restriction enzymes are named from the bacterium from which it was discovered. For example: __________________
- ______ – the first letter of the ___________ name of the bacteria (Escherichia)
- ______ – the first two letters of the ____________ name of the bacteria (coli)
- ______ – a particular _________________ of this bacteria (strain RY13)
- ______ – the particular ________________ among several produced by this strain
- Restriction enzymes cut DNA in areas of specific base pair sequences, called ____________________
- In general, a restriction site is a 4- or 6-base-pair sequence that is a ______________________
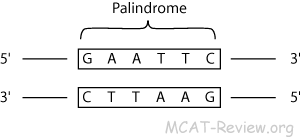
- Each different restriction enzyme (and there are hundreds) has its own restriction site. The ends of the DNA produced after being cut with a restriction enzyme can be either “blunt” or “sticky”
Restriction Enzyme | Restriction site | Sticky or Blunt |
| Restriction Enzyme | Restriction site | Sticky or Blunt |
EcoRI | 5’ GAATTC 3’ 3’ CTTAAG 3’ |
|
| AluI | 5’ AGCT 3’ 3’ TCGA 5’ |
|
BamHI | 5’ GGATCC 3’ 3’ CCTAGG 5’ |
|
| SmaI | 5’ CCCGGG 3’ 3’ GGGCCC 5’ |
|
HindIII | 5’ AAGCTT 3’ 3’ TTCGAA 5’ |
|
| HhalI | 5’ GCGC 3’ 3’ CGCG 5’ |
|
Electrophoresis
Gel Electrophoresis is the most widely used technique to separate pieces of DNA according to their size
View the animation at http://www.sumanasinc.com/webcontent/animations/content/gelelectrophoresis.html and answer the following questions:
What is the purpose of the restriction enzymes? If the enzymes were not added, how many bands of DNA would appear on the electrophoresis gel?
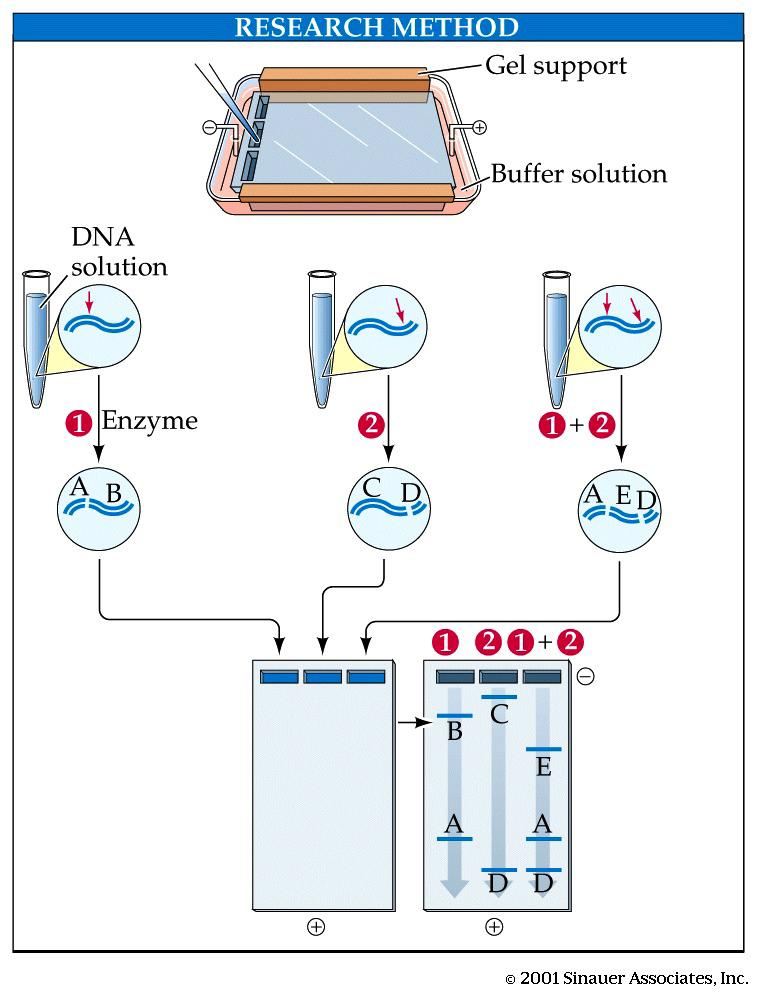
What is the purpose of the agarose gel?
What is the purpose of adding blue “tracking” dye to the DNA samples?
Explain why DNA has an overall negative charge.
Why is the fact that DNA has a negative charge so important in the gel electrophoresis process?
Explain how an agarose gel can separate DNA fragments of different lengths, including a description of which size molecules move furthest in the gel.
In one or two sentences, summarize the technique of gel electrophoresis.
Basics behind DNA Profiling
- Although 99.9% of human DNA sequences are the same in every person, enough of the DNA is different that it is possible to distinguish one individual from another, unless they are monozygotic ("identical") twins.
- DNA profiling uses repetitive ("repeat") sequences that are highly variable, called variable number tandem repeats (VNTRs).

- VNTR loci are very similar between closely related humans, but are so variable that unrelated individuals are extremely unlikely to have the same VNTRs.
- The different sizes of the DNA that result from different numbers of VNTRs will appear as different sizes of DNA bands in electrophoresis.
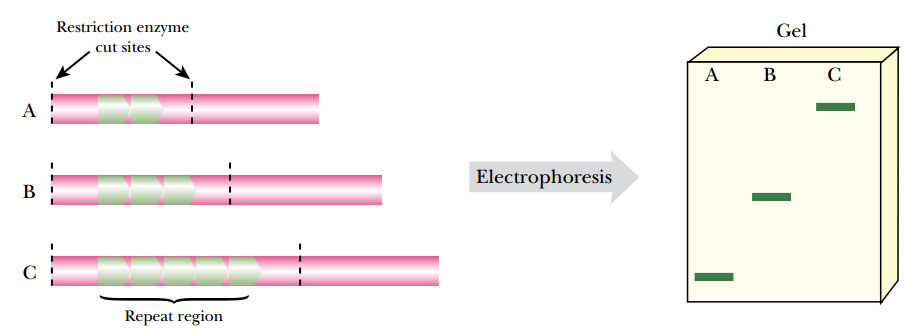
- DNA can be analyzed to:
- Samples can be obtained from:
